
Goal
The basic goal of our project is to fabricate a device aiming to regulate a physiological process and help it recover to a stable state. The detailed steps are introduced as follows:
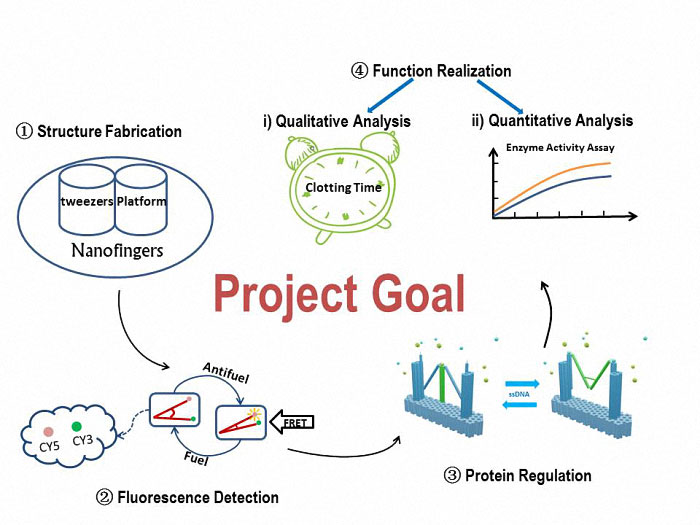
Figure 1. A simple view of our goal and experiment.
To construct our device, Nanofingers, we will utilize the software, CADnano, for the structural design. Through an origami experiment which is a slowly anneal process, we can eventually get our device. The correctively assembled Nanofingers will be proved by TEM and AFM.
To verify the form change, we will attach fluorophores (Cy3 and Cy5) to the two arms of tweezers A. The variation of the fluorescent intensity demonstrates that Nanofingers functions well to change its form in response to an ssDNA signal.
To check the function of capturing a protein, we will compare the two conditions, which are with and without a captured protein on Nanofingers. An electrophoresis is adopted and there will be an obvious difference in migration speed between the two conditions, suggesting Nanofingers successfully captures our target proteins.
To reveal the influence Nanofingers exerts to the proteins, we will analyze the function qualitatively and quantitatively to give a vivid view on its effectiveness.
To prove that Nanofingers can actually reduce the activity of the captured protein. We will measure the clotting time since the protein we use in practice is thrombin which plays a role in blood coagulation. The time is expected to be increased due to the appearance of Nanofingers.
Enzyme activity assay is conducted to quantitatively indicate the variation of the enzyme’s activity.

Design
Contents
As is illustrated in Figure1, Nanofingers is devised to regulate two related proteins in vivo. The homeostasis requires the organism to control a physiological process at least from two aspects, promoting it and inhibiting it. Regulation from these two aspects simultaneously can lead to an amplified influence on the process. When we get injured, a signal will be released and identified by Nanofingers. Triggered by the signal, Nanofingers starts to release proteins helping to coagulate blood and through a linkage DNA strand, the nearby structures change their conformation to capture proteins involved in blood dissolution. The proteins we use are thrombin and hirudin respectively. The efficiency of blood coagulation is highly increased and the organism can soon recover to its homeostasis. The basic structures of our device are introduced as follows.
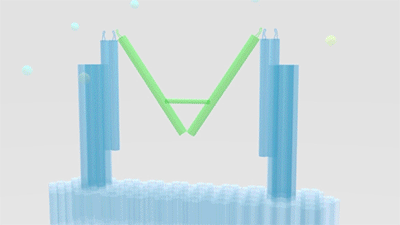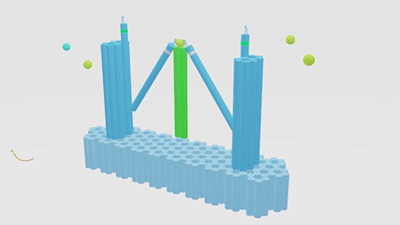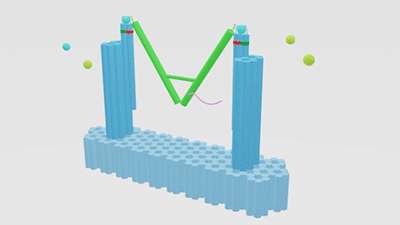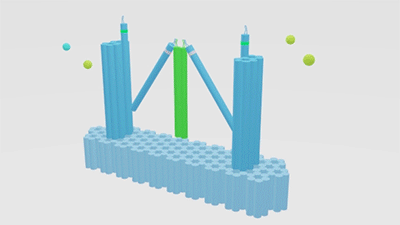
Figure 1. Nanofingers regulates “Pairs of proteins”.
The main constituents of our device are three tweezer-like structures which can be categorized into two types, tweezer A and tweezer B (figure 2c). Two DNA double crossover (DX) motifs are joined by a Holliday junction to form a tweezer-like DNA machine (figure 2a). Two kinds of aptamers are linked at the end of it to specifically bind two different parts of a protein. When the two aptamers are placed in a close distance, they show a cooperative effect, resulting in a significant enhancement of binding affinity and specificity to the protein. A DNA motor is incorporated into the middle of a tweezer, which can cycle between stem-loop and double helix structures driven by a strand-displacement reaction and it works to adjust the distance between the arms of these DNA tweezers. Two different ssDNA/ssRNA involved in the cycle are considered as two kinds of signal and can trigger the form change of the tweezers. Two aptamers hence approach with each other and become able to capture the target protein. In contrary, when the two aptamers are separated, the protein will be released (figure 2b). The biggest differences between the two types of tweezers are that tweezer B is equipped with a DNA motor but has no aptamer while tweezer A has aptamers without DNA motor (figure2c). According to our design, tweezer B is regarded as an activator and tweezers A are responders. Nanofingers is the machine that brings them together.
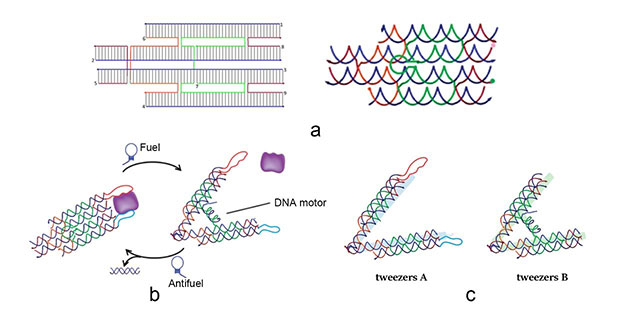
Figure 2. a (Left): The design of DNA tweezers. a (right): The Holiday junction of tweezers. b: Schematic illustration of reversible regulation of target binding affinity by the DNA tweezers. c: There are two types of tweezers, A and B. Here we show the differences between them.
As designed, the open and closed conformation of the DNA machine should induce a significant change in the distance between the ends of the two arms, and this can be verified by Forster resonance energy transfer (FRET). To demonstrate that tweezer B can be activated by signal “fuel & antifuel” and change the conformation of tweezers A, we add two fluorophores to the two arms of tweezers A. They can show different fluorescence intensity in the two states, which is caused by FRET. As shown in Figure 3, the two arms of tweezers A are modified with fluorophores at their ends, one with Cy3 (donor) and the other with Cy5 (acceptor). Besides, the binding sites of fluorophores have been accurately calculated to lead to an efficient FRET when tweezers A are in the closed state.
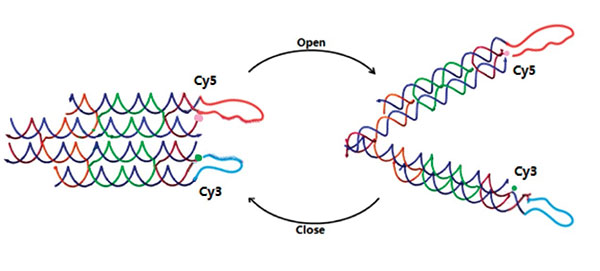
Figure 3. DNA tweezers A with fluorophores.
In the real wet lab experiment, we choose bovine thrombin as the model object to demonstrate the ability of our DNA machine to regulate two related proteins. According to the literature, aptamer A (apt-A: 29mer, 5’-AGT CCG TGG TAG GGC AGG TTG GGG TGA CT-3’) and aptamer B (apt-B: 15mer, 5’-GGT TGG TGT GGT TGG-3’) can be used in this tweezer-like structure and achieve the function as expected. So when the tweezer is in the closed state, two aptamers will be kept 4-6 nm apart as designed, and with a distance-dependent bivalent interaction, the tweezer can strongly bind thrombin. When in the open state, this interaction will disappear in terms of the distance between aptamers, and thrombin will be released (figure 4). We design the two aptamers for tweezers A to capture and release thrombin. Unfortunately, there are no aptamer records about hirudin, but it’s enough to demonstrate the feasibility of our project by just applying thrombin.
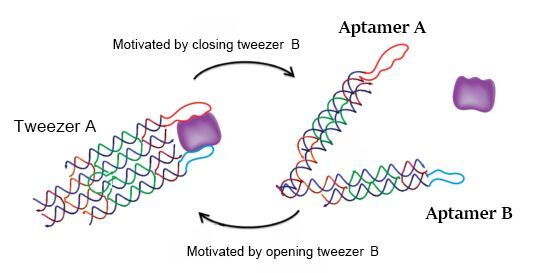
Figure 4. Reversible regulation of target binding affinity by the DNA tweezers
Inspired by the idea that managing different kinds of proteins can sustain physiological homeostasis to the utmost, here we design a linkage structure between each pair of tweezers which can regulate different proteins separately. To take full advantage of the tweezer-like structure, a DNA strand is used to link the two arms of the different tweezers. Tweezers A are specifically bound to thrombin. At the same time, the most noticeable difference between tweezers A and tweezer B is that, tweezer B can be controlled by special DNA signal as “fuel and antifuel” but tweezers A can’t. In this case, in response to the signal, tweezer B can switch from open state to closed state, which leads to an opposite effect of making tweezers A open by the linkage. So, in this process, different types of proteins can be regulated by tweezers A and tweezer B simultaneously. In this project, we apply the thrombin. However, we strongly recommend other “Pairs of proteins”, for instance, insulin & glucagon.

Figure 5. DNA origami is designed by cadnano.To match the real shape and give a clearer demonstration of the designed structure, we modified the image since the illustration of structure design in cadnano is restricted and differs from the real shape.
In our project strategies, we have demonstrated that a fixed platform is needed for the tweezers A and tweezer B to keep in different states and provide a switch pivot point. So, we use cadnano for our DNA origami platform design (figure 5), and use the CANDO to test the stability of this structure. The structure can be simplified as the model, in which the tweezers are ranked as “ABA”. Meanwhile, to achieve a high stability and keep in correct conformation, one arm of tweezers A is made up of a part of the fixed platform (figure 6).
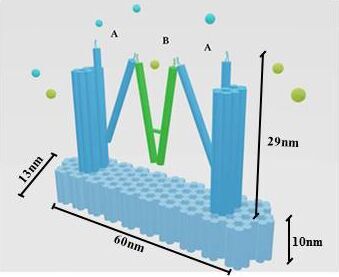
Figure 6. A fixed DNA platform stabilizes the tweezers. The tweezer in green is tweezer B, and tweezers A stand at two sides.The dimension of Nanofingers is labeled in the image.
With the function of regulating different proteins, Nanofingers has been demonstrated that it can be applied to regulate the concentration of thrombin to keep our body in homeostasis. There are some reactions we can take advantages of, as a specific DNA or RNA signal can be produced when we are bleeding. And in response to this signal, Nanofingers can change its conformation to operate automatically to release thrombin, which accelerates the blood coagulation (Figure 7). Once the coagulation is finished, Nanofingers will capture thrombin in response to another specific DNA or RNA signal, and it is a good way to avoid thrombus.
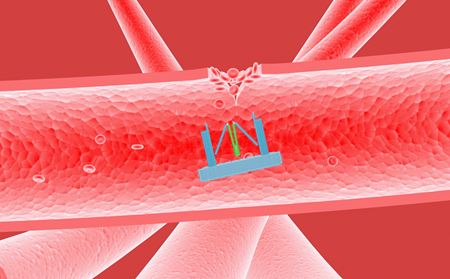
Figure 7. Nanofingers releases thrombin and helps accelerate blood coagulation
In this regulation system of Nanofingers, we can simultaneously regulate a physiological process from two aspects and remarkably enhance the regulation ability. Since we adjust the concentration of three proteins with one signal by our nanofingers, it means a lot to regulate the metabolic reaction more effectively.
We also want to explain this strategy from a very logical aspect. Here we employ the general system theory to demonstrate the vast application prospect of Nanofingers (Figure 8). As we have referred, insulin & glucagon can also be used.
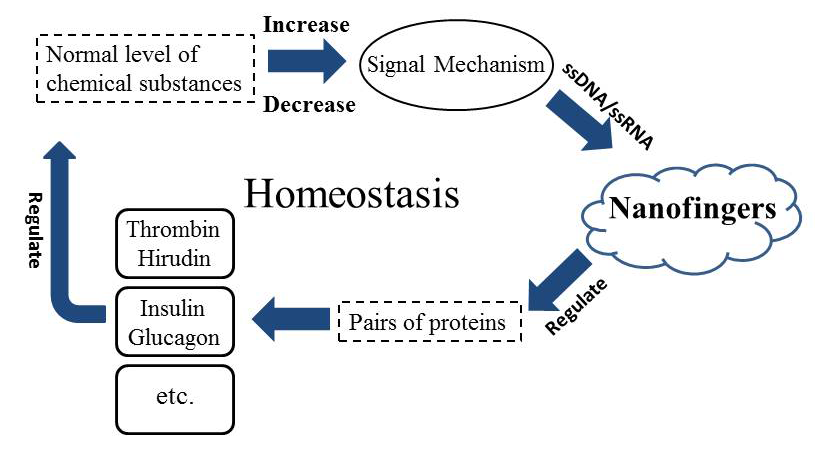
Figure 8. The general system theory illustrates the important role of Nanofingers in sustaining the homeostasis.
In the course of designing the structure of Nanofingers, we noticed that it features like the Vulcan salute when tweezers A are closed and tweezer B is open. The Vulcan salute is shown as double-fingered version of victory sign, which originates from Star Trek with the meaning of blessing for "Live long and prosper". Nanofingers is our Vulcan salute to all the world, expressing the best wishes “Live long and prosper”.
